Example 2: Bacteria Genomes Alignment
Data download
There are two perl programs provided to help users to download genome sequences from NCBI conveniently. The basic syntax are as shown below.
Get_seq.pl is used for downloading individual genome sequence.
perl ~/Scripts/withncbi/util/batch_get_seq.pl \$genome_id $dir
Batch_get_seq.pl is used for downloading a batch of genome sequences at a time.
perl ~/Scripts/withncbi/util/batch_get_seq.pl -r -p -f yeast_name_seq.csv 2>&1 | tee yeast_name_seq.log
The program scripts and detailed syntax are here.
Genome sequences downloaded from NCBI will be processed to adjust the formats of sequence headers automatically, so users need not handle it manually. Downloaded genomes are named as the corresponding RefSeq identities in NCBI, which will also be processed as the genome name in result files.
We also provided a test account stored with the data used for example tests. You can log into the account for bacteria with the username "bacteria" and password "password".
The process of alignment is similar to that of yeast. You can skip this part if you have read the instruction for yeast. If not, the instruction below will help you get a hands on start.
Upload
click on "Upload" to enter the page after you log in the site. The upload page shows an upload dialog.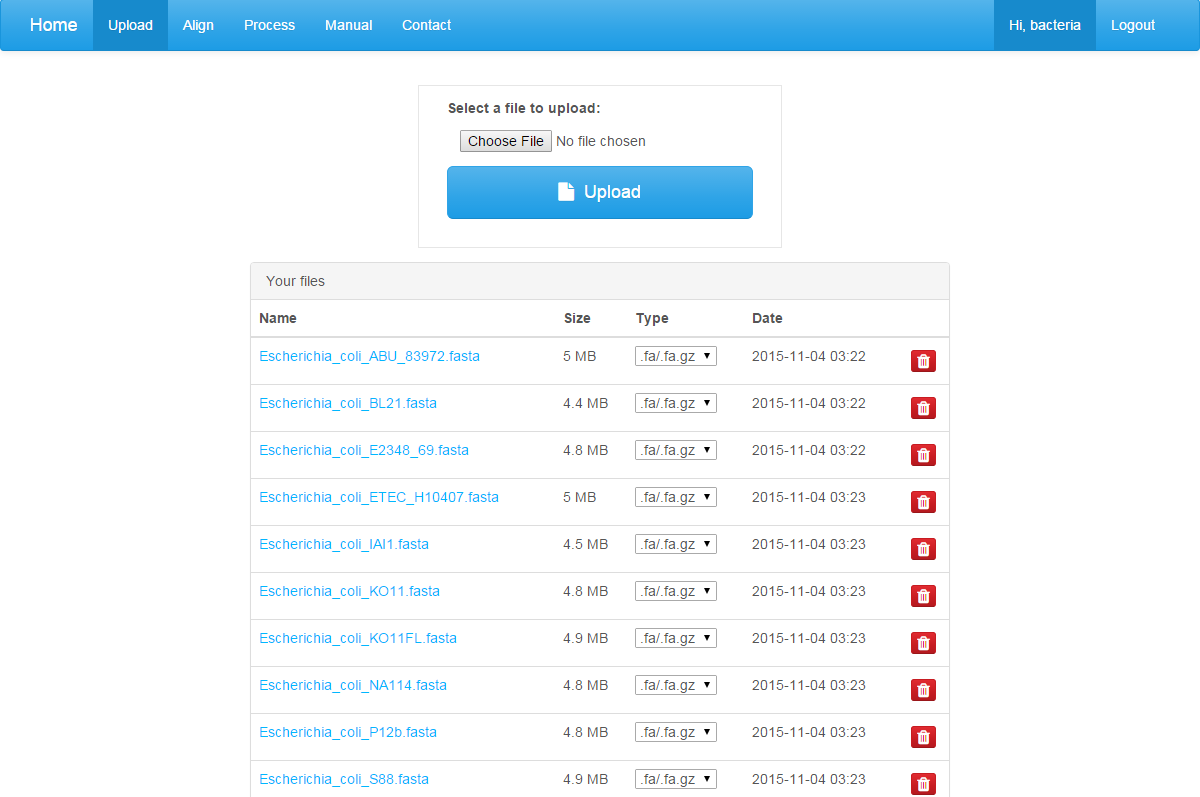
If you need to upload a genome file, you can select a file in the open dialog box, and click on the “Upload” button. For a web-based alignment instead of process through a virtual machine, the individual file size is limited to 30M. Only three kinds of file types are accepted: .fasta, .fasta.gz, .newick. You can click on the red button to remove an uploaded file. Do not refresh the page while uploading a file!
Alignment
You can click on the “Align” button on the menu to enter the align page.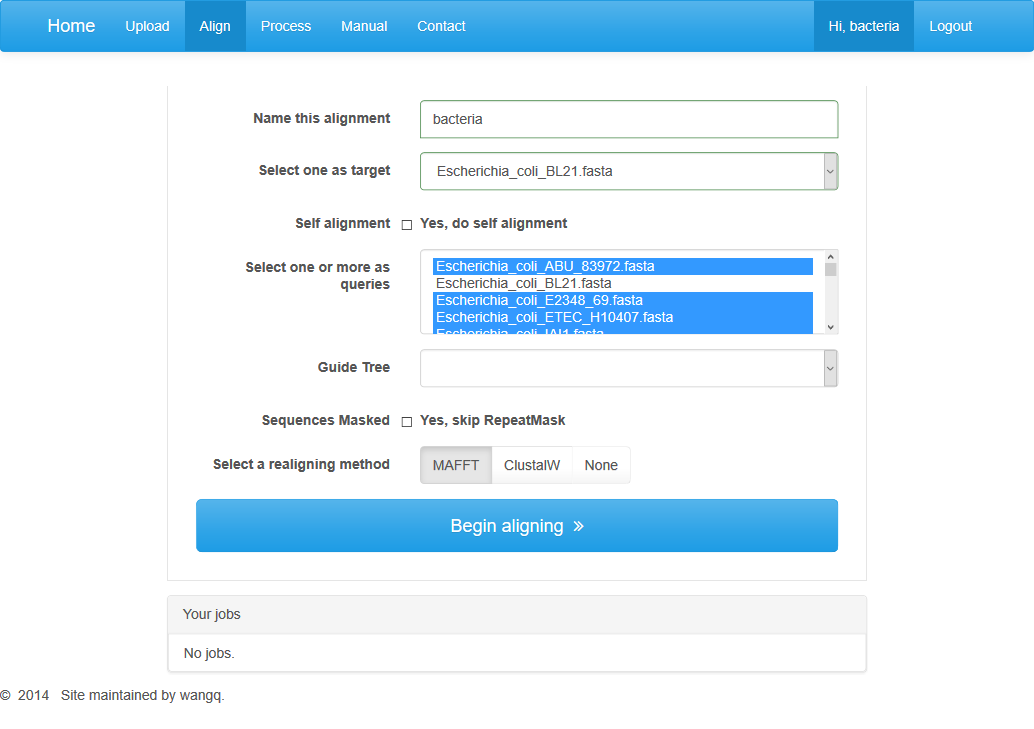
At this page you should set some parameters for the construction of alignments, such as the name of this alignment job, a target genome, at least one query genome. Besides, you can choose to provide a guide tree or not. The absence of a guide tree will take a while for us to generate one. We also provide a recommended realigning method.
After parameter settings are completed, you can click on “Begin aligning” button to jump to the processing page automatically.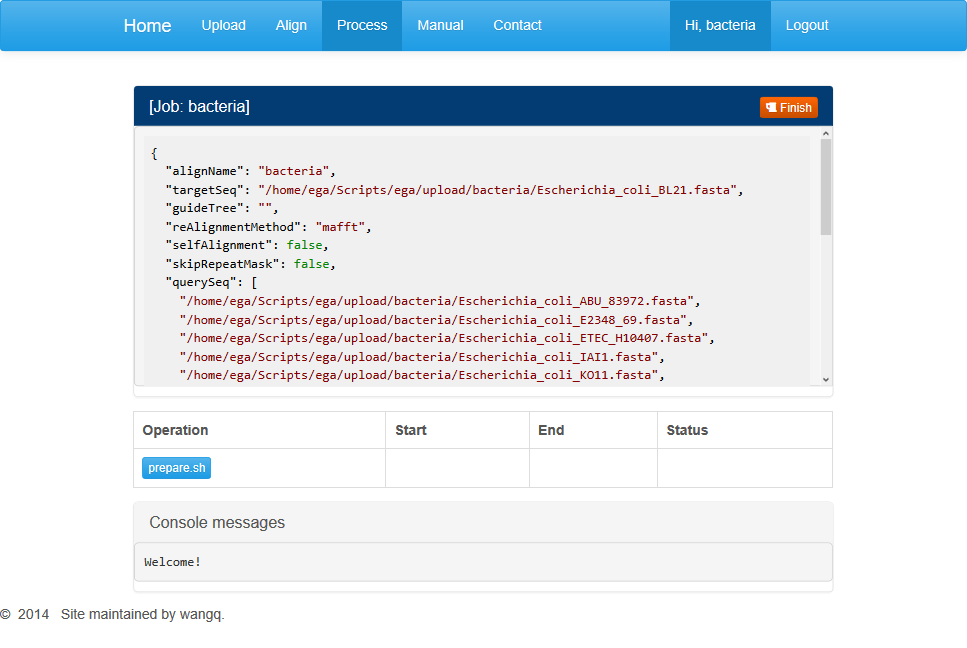
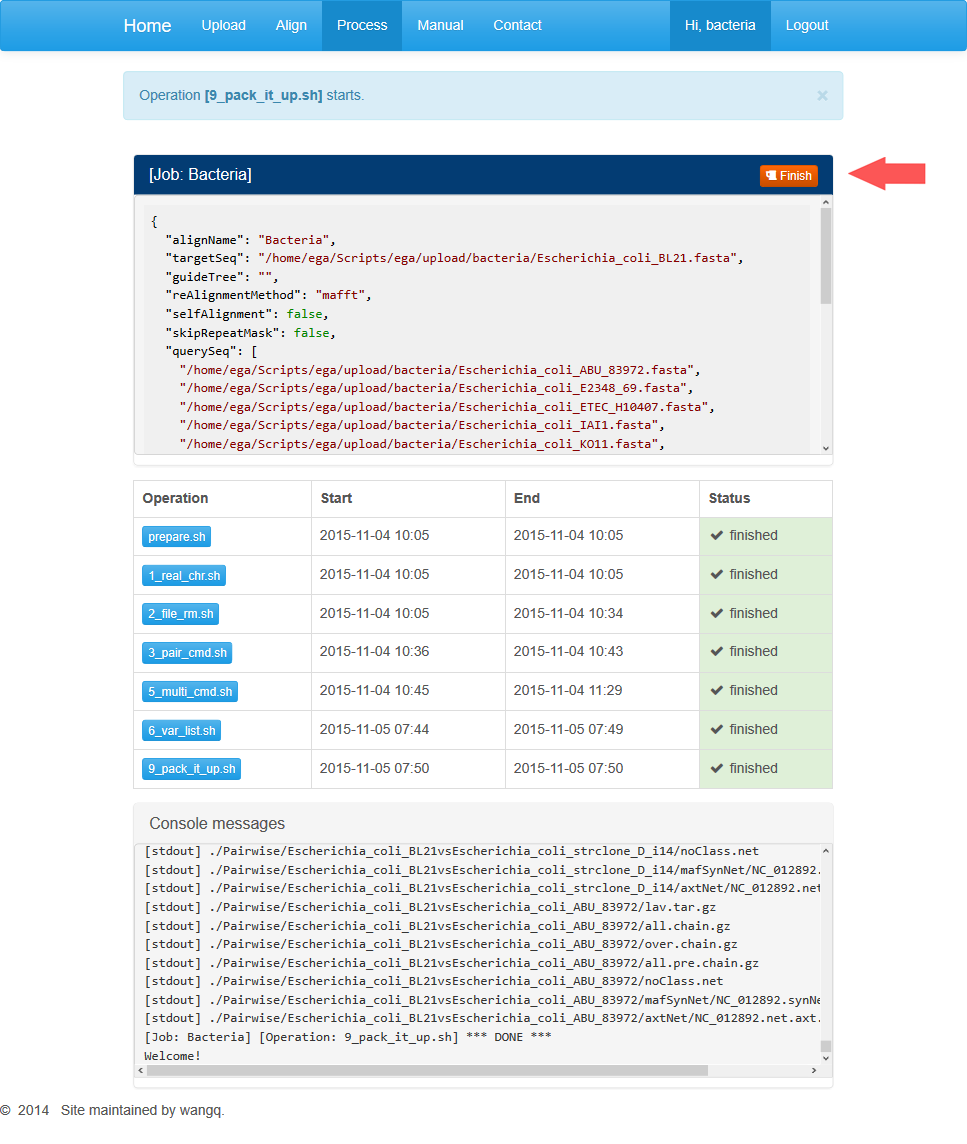
This page displays three dialog boxes. The dialog above contains parameters set in the previous step. You can’t open a new job if the current one has not finished. When your job is completed or you want this job terminated, you can click on the red button at the top-right corner and open up a new task. Notice that terminated job can’t recover.
The middle dialog contains buttons for different processing stages. Among them, the first step is data pre-processing. You can click on the “prepare.sh” to start a job running.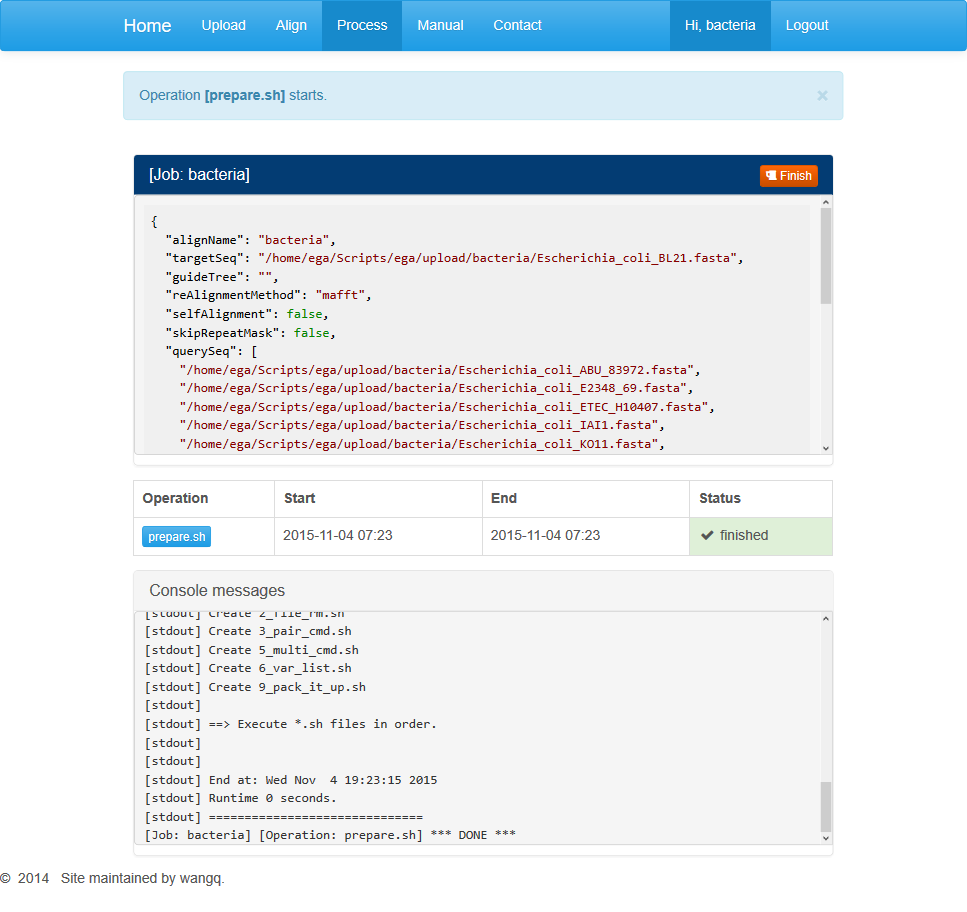
The information section contains general information about the running task including the start and end times and running status.
Below is progress information for the particular run step. When the last line shows “*DONE*”, the step is supposed to be completed. Users may refresh this page and move on to the next step.
After the pre-processing completes, refresh the page and buttons for the rest steps will appear in the middle dialog box. Users may click on buttons in sequence to generate an alignment. For every step, when the last line shows “*DONE*”, the task is completed and you can move on to the next step.
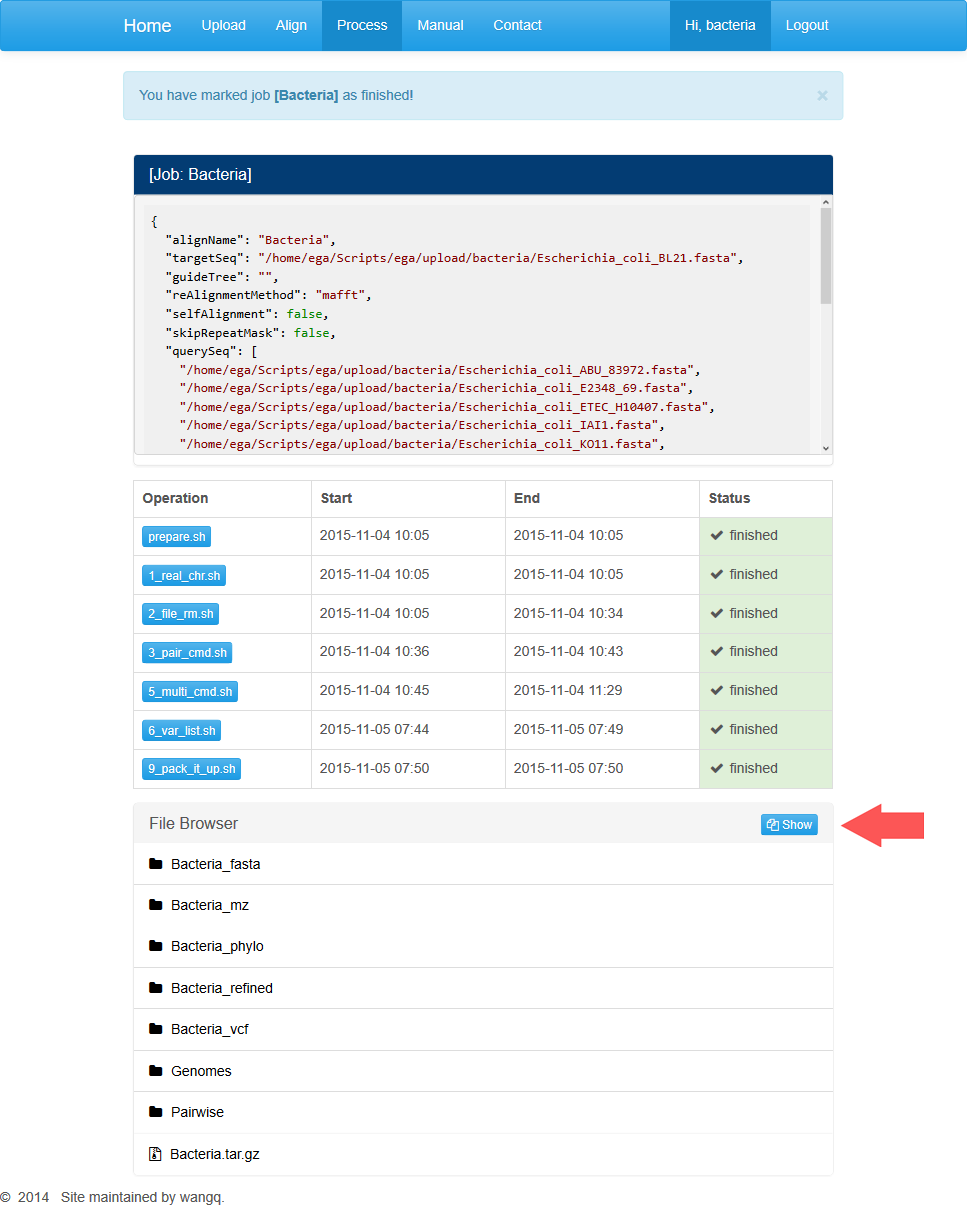
Downloading results
After all jobs are done, users may click on the red “finish” button and download the results generated in every step.