For multiple genomes alignment
The result folder contains folders listed below: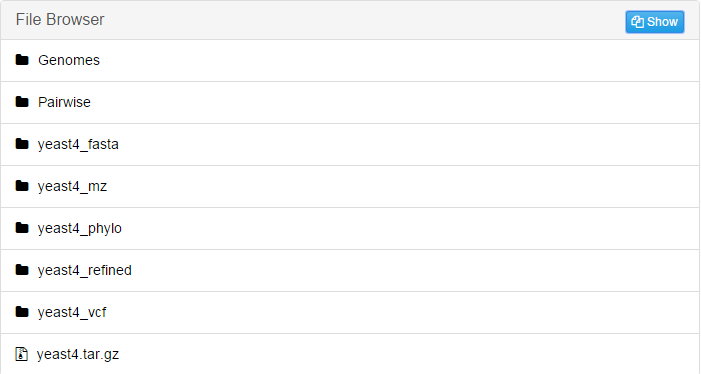
Genomes: Contains the information of individual files processed in this alignment, including: .fasta file after data preparation, information after repeat mask and the size analysis of the genome.
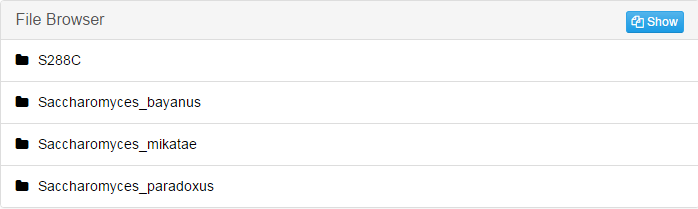
Take "S288c" folder as an example: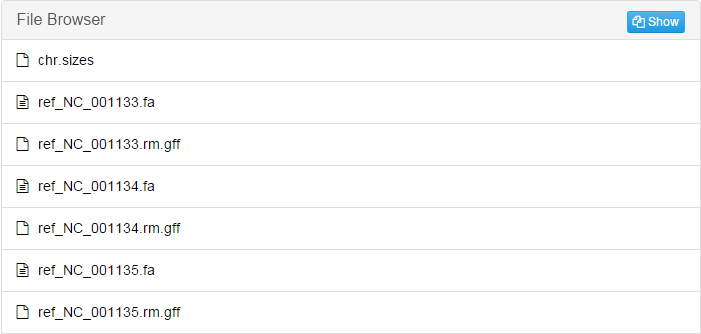
If your species has more than one chromosomes, there will be corresponding files for each of them.Pairwise: Contains the results of pairwise alignment between reference genome and every other genomes in this alignment.
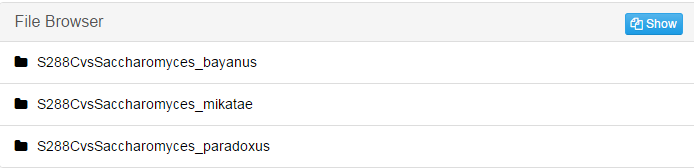
Take "S288CvsSaccharomyces_bayanus" folder as an example: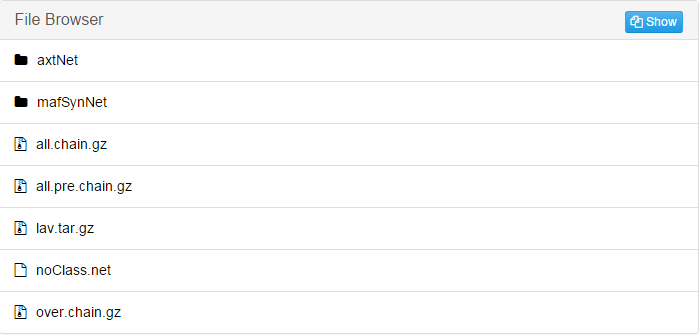
jobID*_mz: Combine the individual result file of pairwise alignment into .maf file by Multiz.
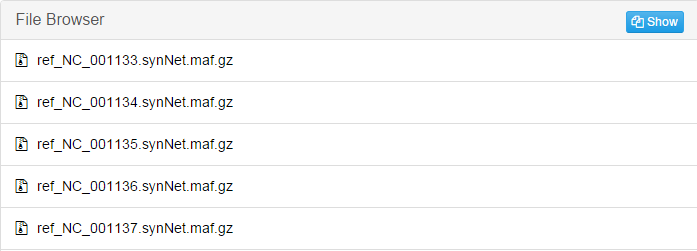
This screenshot is only part of the result. There will be a file for each individual chromosome in reference genome.jobID_fasta: Convert .maf file produced in the former step into .fasta file.
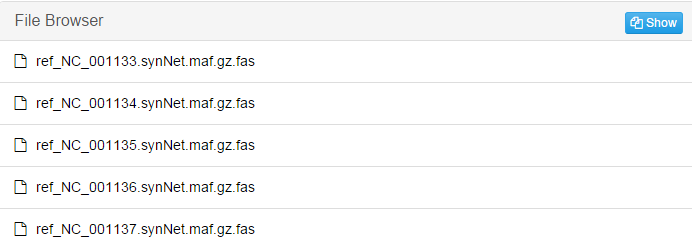
This screenshot is only part of the result. There will be a file for each individual chromosome in reference genome.jobID_refined: The .fasta file in the former step may contain gaps. After refinement, the final result of alignment is stored in the refined .fasta file.
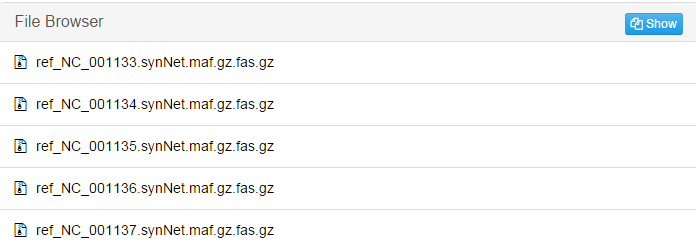
This screenshot is only part of the result. There will be a file for each individual chromosome in reference genome.jobID_vcf: Contains a .vcf file stored with the statistical analysis of SNP and indel information in the alignment results.
This screenshot is only part of the result. There will be a file for each individual chromosome in reference genome.jobID_phylo: Contains the result files related to the phylogenetic tree, including: the final phylogenetic tree in .nwk and .phy file style, a vector graphic in .svg file style and other intermediate files produced during the construction process by RAxML.
*jobID refers to the parameter you set for the name of the alignment.